Recombination Operators: Permutation, Integer, and Real-Valued Crossover
We have been introduced to recombination operators before; however, that was merely an introduction. There are dozens of different Evolutionary Algorithm recombination operators for any established genotype; some are simple, some are complicated.
For a genotype representation that is a permutation (such as a vector[1], bit-string, or hash-map[2]), we have seen a possible recombination operator. Our 3-SAT solver uses a very popular recombination technique: uniform crossover.
Furthermore, we know a permutation is not the only, valid genotype for an individual: other possibilities can include an integer or a real-valued number.
Note, for simplicity, we will discuss recombination to form one offspring. This exact process can be applied to form a second child (generally with the parent's role reversed). Recombination can also be applied to more than two parents (depending on the operator). Again, for simplicity, we choose to omit it[3].
First, let us start with permutations.
Permutation Crossover
In regard to premutation crossover, there are three, common operators:
- Uniform Crossover
- $N$ -Point Crossover
- Davis Crossover
Uniform crossover we have seen before. We consider individual elements in the permutation, and choose one with a random, equal probability. For large enough genotypes, the offspring genotype should consist of 50% of the genotype from parent one, and 50% of the genotype from parent two.
$N$-Point crossover considers segments of a genotype, apposed to individual elements. This operator splits the genotype of Parent 1 and Parent 2 $N$ times (hence the name $N$-point), and creates a genotype by alternating segments from the two parents. For every $N$, there will be $N + 1$ segments. For 1-point crossover, the genotype should be split into two segments, and the offspring genotype should be composed of one segment from Parent 1, and one segment from Parent 2. For 2-point crossover, there will be three segments, and the offspring genotype will have two parts from Parent 1 and one part from Parent 2 (or two parts, Parent 1, one part, Parent 2).
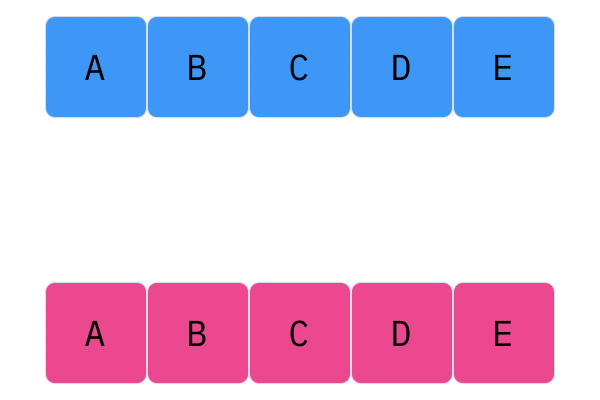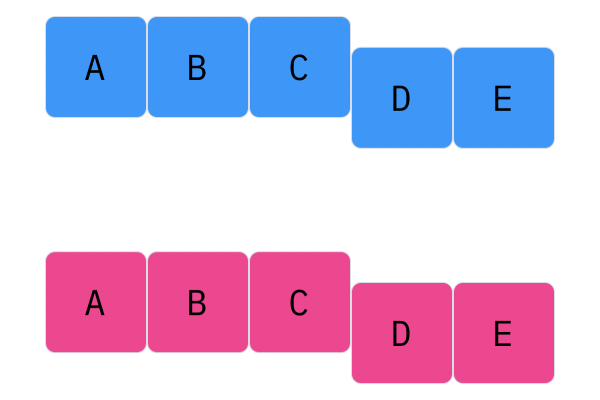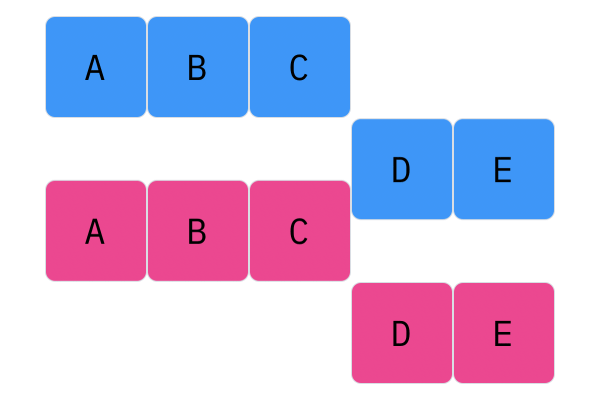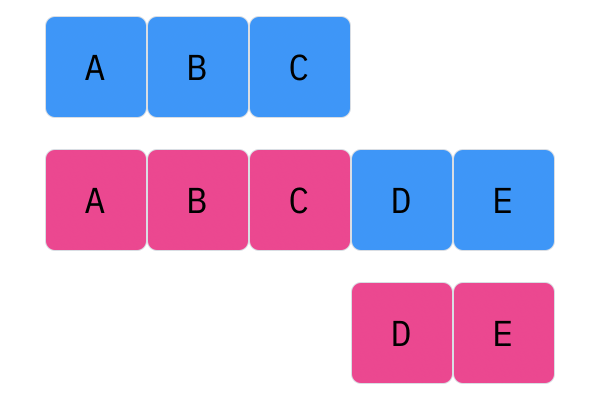
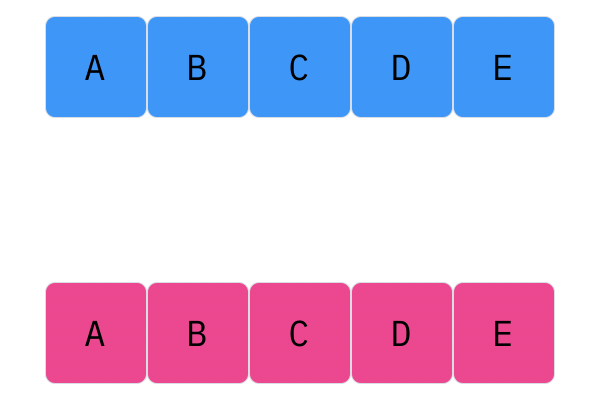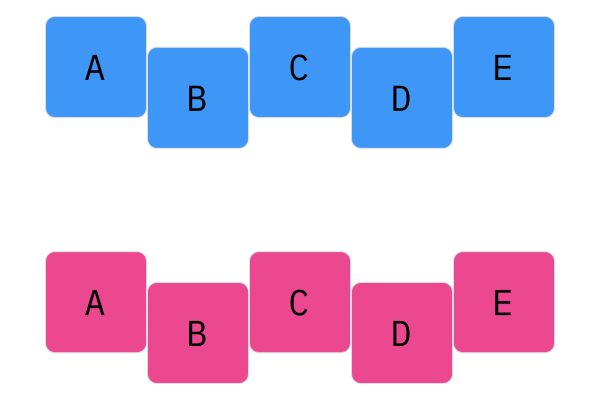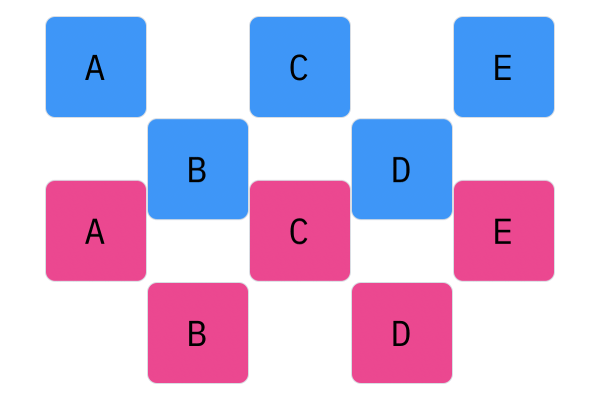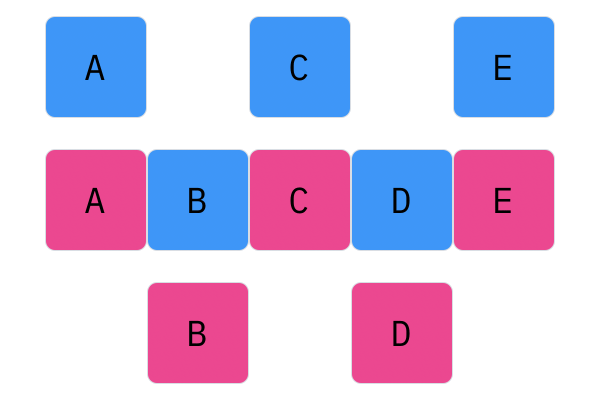
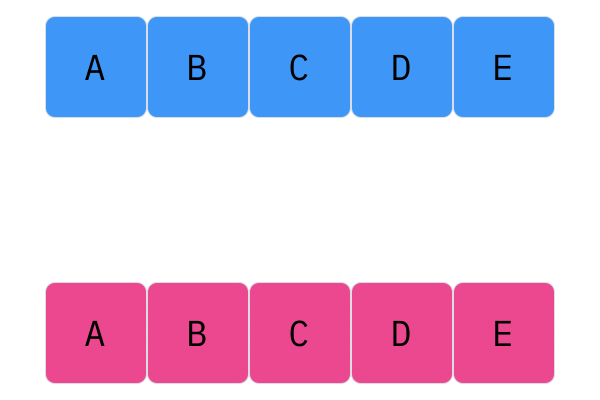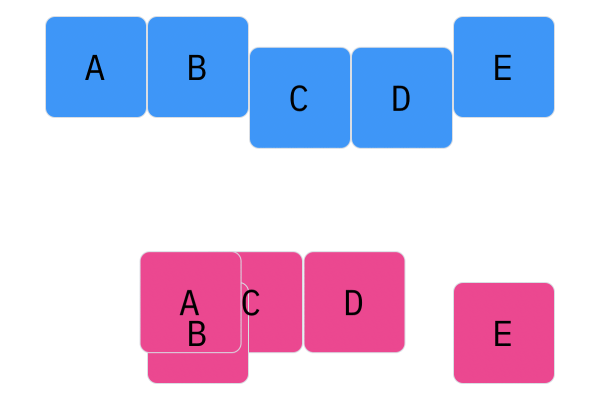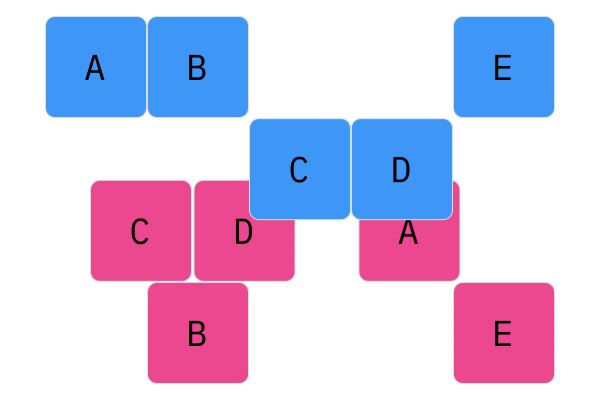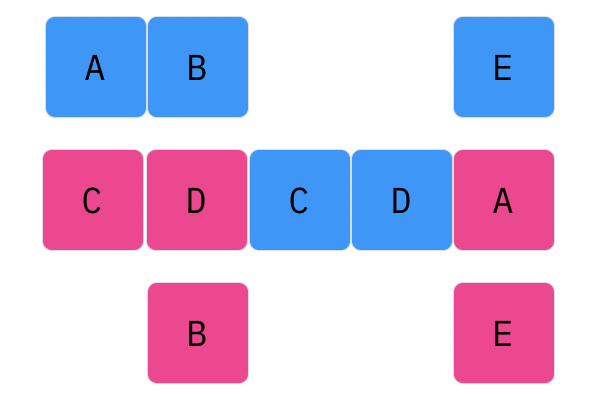
Davis Crossover tries to preserve the ordering of the genotype in the offspring (apposed to the previous methods, where ordering was not considered). The premise is a bit complicated, but bear with me. Pick two random indices ($k_1$ and $k_2$), and copy the genetic material of Parent 1 from $k_1$ to $k_2$ into the offspring at $k_1$ to $k_2$. Put Parent 1 to the side, his role is finished. Start copying the genotype of Parent 2 starting at $k_1$ to $k_2$ at the beginning of the offspring. When $k_2$ is reached in the parent, start copying the beginning of Parent 2 into the genotype, and when $k_1$ is reached in the parent, skip to $k_2$. When $k_1$ is reached in the offspring, skip to $k_2$, and start copying until the end. If this seems a complicated (it very much is), reference the accompanying figure.
Those are considered the three, most popular choices for permutations. Now, let us look at integer crossover.
Integer Crossover
Integer crossover is actually quite an interesting case; integers can be recombined as permutations or real-valued numbers.
An integer is already a permutation, just not at first glance: binary. The individual bits in a binary string are analogous to elements in a vector, and the whole collection is a vector. Now it is a valid permutation. We can apply uniform crossover, $N$-point crossover, or Davis Crossover, just as we have seen.
An integer is also already a real-valued number, so we can treat it as such. Let's take a look at how to recombine it.
Real-Valued Crossover
Real-Valued crossover is different than methods we have seen before. We could turn it into binary, but that would be a nightmare to deal with. However, we can exploit the arithmetic properties of real-valued numbers — with a weighted, arithmetic mean. For a child (of real value) $z$, we can generate it from Parent 1 $x$ and Parent 2 $y$ as such:
$$
z = \alpha \cdot x + (1 - \alpha) \cdot y
$$
Now, if we want to crossover a permutation of Parent 1 and Parent 2, we can do so for every element.
$$
z_i = \alpha \cdot x_i + (1 - \alpha) \cdot y_i
$$
This can be shown to have better performance than crossover methods discussed, but would entirely depend on use case.
Implementing Permutation Recombination
As always, we will now tackle implementing the permutation crossovers we've had before. None of them are incredibly complicated, except possibly $N$-point crossover.
class Individual
...
@staticmethod
def __uniform_crossover(parent_one, parent_two):
new_genotype = SAT(Individual.cnf_filename)
for variable in parent_one.genotype.variables:
gene = choice([parent_one.genotype[variable], parent_two.genotype[variable]])
new_genotype[variable] = gene
individual = Individual()
individual.genotype = new_genotype
return individual
@staticmethod
def __n_point_crossover(parent_one, parent_two, n):
new_genotype = SAT(Individual.cnf_filename)
variables = sorted(parent_one.genotype.variables)
splits = [(i * len(variables) // (n + 1)) for i in range(1, n + 2)]
i = 0
for index, split in enumerate(splits):
for variable_index in range(i, split):
gene = parent_one.genotype[variables[i]] if index % 2 == 0 else parent_two.genotype[variables[i]]
new_genotype[variables[i]] = gene
i += 1
individual = Individual()
individual.genotype = new_genotype
return individual
@staticmethod
def __davis_crossover(parent_one, parent_two):
new_genotype = SAT(Individual.cnf_filename)
variables = sorted(parent_one.genotype.variables)
split_one, split_two = sorted(sample(range(len(variables)), 2))
for variable in variables[:split_one]:
new_genotype[variable] = parent_two.genotype[variable]
for variable in variables[split_one:split_two]:
new_genotype[variable] = parent_one.genotype[variable]
for variable in variables[split_two:]:
new_genotype[variable] = parent_two.genotype[variable]
individual = Individual()
individual.genotype = new_genotype
return individual
Recombination In General
By no means is recombination easy. It took evolution hundreds of thousands of years to formulate ours. The particular permutation operator to use entirely dependent on the context of the problem; and most of the time, it is not obvious by any stretch. Sometimes, there might not even be an established crossover operator for a particular genotype.
Sometimes, you might have to get a little creative.
